GPU accelerated molecular dynamics
YASARA's molecular dynamics algorithms[1] can now be accelerated using GPUs from AMD, nVIDIA and Intel, in Linux, Windows and MacOS. The actual performance is shown on the benchmark page.
YASARA uses the GPU to calculate the non-bonded interactions (Van der Waals and real-space Coulomb forces), all the rest (PME, bonded intraactions, NMR restraints…) is done by the CPU. This approach has a few advantages:
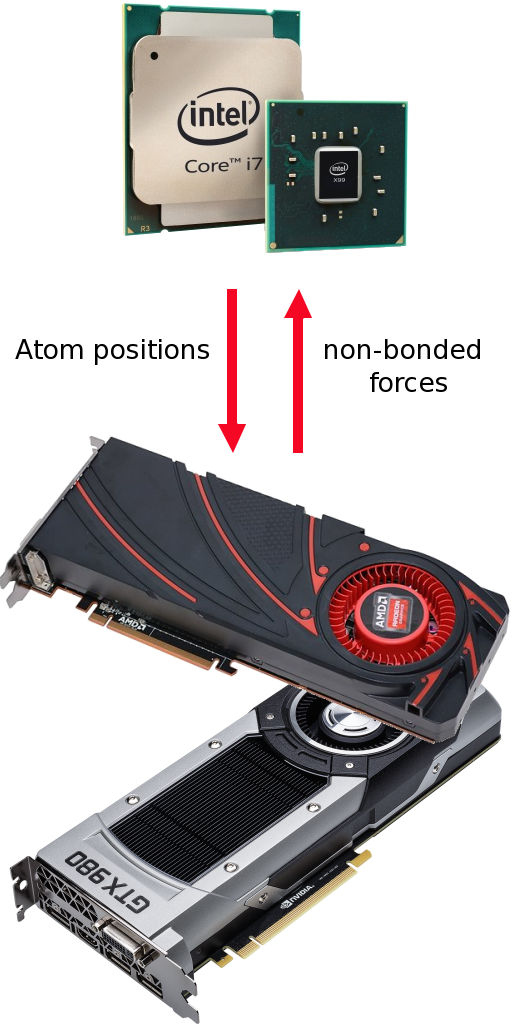
- The CPU's power is not wasted by letting it run idle, especially since some tasks are better handled by the CPU (the GPU architecture is quite different).
- Complicated algorithms developed over the past decades don't have to be rewritten for the GPU and are immediately available (knowledge- based force fields for protein refinement, NMR restraints…)
- Macros that interact with the simulation (steered MD etc.) work unchanged.
The only real disadvantage is that you cannot upgrade an old computer by inserting a top graphics card. Instead, the power of the CPU must match the GPU (or the GPU will run idle) - but having a fast CPU for everyday work is not really a disadvantage at all.
Recommended CPUs:
To keep the GPU busy, a fast CPU is required. Depending on your budget, we recommend most recent CPUs from AMD like the new Ryzen 9000 series released in 2024. Intel has unfortunately fallen far behind due to problems with die shrinks in their plants. To match AMD's core count, Intel CPUs now include many 'efficiency cores', which are unfortunately so slow that they likely have to be disabled in the BIOS, e.g. the Intel Core i9-14900HX has 24 cores, of which only 8 can be used in practice for high performance MD. Another thing to keep in mind is that CPU clock speed tends to be more important than CPU core count. For example Intel's giant Xeons with huge caches and countless cores are less attractive, because they combine a significantly higher price tag with a low clock frequency, that is even throttled further down when using high performance AVX code (like YASARA does). For the same reason, machines with two or more CPU sockets cannot be recommended. The cores should all have equally fast access to memory, that's why we don't recommend Ryzen Threadripper 2990WX and 2970X, where you have to fiddle with extra tools to tune memory access. Also note that the more threads a CPU can execute, the larger the simulated system must be. The very small DHFR benchmark with 23786 atoms can keep 16 threads busy. A PC should have minimally 8 GB RAM to run YASARA smoothly.
YASARA uses the industry standard 'open compute language' (OpenCL) to communicate with the GPU. OpenCL is supported by all major GPU vendors. To estimate how well a certain card from nVIDIA or AMD is suited for MD simulation with YASARA, please visit the CompuBench 1.5 OpenCL benchmark page and look at "Particle simulation 64k".
Since YASARA uses both CPU and GPU at the same time, their power must match. It doesn't make sense to pair an ultra-high end GPU with a low end CPU, because the GPU would spend most of its time waiting. Likewise, a slow GPU as found in notebooks will likely slow down the simulation. If you go for a GPU from nVIDIA, pick a Geforce RTX or newer, it has features that are helpful for YASARA's new Vulkan graphics engine. To estimate performance, look at the FP32 TFLOPs and be careful since the success of AI has led to the development of very expensive GPUs for AI that have neglegible FP32 and thus MD performance (for example the Geforce RTX 4090 has 82 FP32 TFLOPs, while the NVIDIA A100 delivers only 19.5 TFLOPs, i.e. 24%).
Recommended operating systems:- Linux delivers the highest performance, thanks to its frequent use on clusters and the extensive optimization work done to make good use of these expensive resources.
- Windows, being the primary video game platform, has also been well optimized and gets very close to Linux (within a few percent), so the difference is only measurable but not noticeable.
- MacOS has been heavily optimized for power efficiency and long battery life on mobile devices, but not so much for performance. It has difficulties dealing with programs that spawn multiple threads to fully exploit the CPU's potential. As a result, MD simulations of small proteins may run noticeably slower, but the difference becomes smaller with growing protein size.
R E F E R E N C E S
[1] New ways to boost molecular dynamics simulations
Krieger E, Vriend G (2015). J.Comput.Chem. 36, 996-1007, full text








